RISK OF WRONG PRIORITY SCREENING RESULTS
Prenatal screening to detect fetal chromosomal abnormalities has become common among pregnant women. The screening methods are known: fetal ultrasound (nochal translucency), maternal serum screening (β-HCG and PAPP-A) with reported detection rates of 60–90% and the false positive rate is 5%.
If the results of this screening test indicate a high risk of chromosomal abnormalities in the fetus, then an invasive diagnostic test such as chorionic villus sampling (CVS) is recommended at 12–13 weeks of gestation or amniocentesis at 15–16 weeks of gestation. While these tests are valuable diagnostic tools because of their high accuracy, they are associated with a 0.5 to 1.0% risk of miscarriage. Since the discovery of placental-derived free-cell DNA (cfDNA) in the blood of pregnant women, this has been seen as a potential avenue for screening for fetal abnormalities with high accuracy. Highly accurate and completely safe for pregnant women and fetuses.

In 2011, the NIPT test entered clinical practice and is considered the most accurate prenatal screening test, with an abnormal detection rate of more than 99% and a low false positive (FP) rate. (<0.1%) and has been recommended by many professional associations around the world such as: International Society of Obstetrics and Gynecology (ISUOG) and American Society of Obstetricians and Gynecologists (ACOG), American Society of Obstetrics and Gynecology International Prenatal Diagnosis (ISPD) and the Royal College of Obstetricians and Gynecologists (RCOG).
However, the accuracy of NIPT is also highly dependent on technical factors and many other biological mechanisms. The CfDNA used in the NIPT assay is released from placental trophoblasts rather than directly from the fetus, and genetic differences between the placenta and the fetus can lead to these results. Inaccuracies, such as false-positive or false-negative results, expose pregnancy management to some risk.
False-Negative RESULTS IN THE NIPT TEST
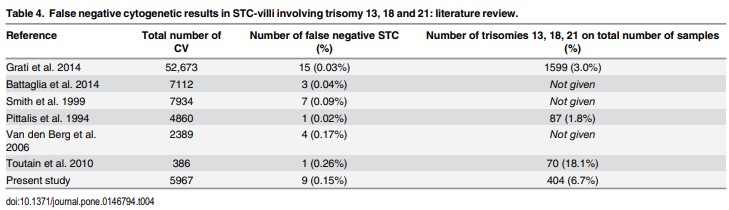
A false negative result in the NIPT test is interpreted as not detecting abnormalities in the fetus but in fact the fetus has abnormalities on the syndromes screened in NIPT. False negative results in NIPT are rare, but there have been many studies around the world reporting on this case. According to the 2020 statistics, the false negative rate in NIPT can range from 0.02% to 0.26% (Figure 1).
Specific causes that can lead to false-negative results in NIPT have been investigated and reported, and the causes are known to be: presence of fetal mosaicism; low fetal fraction (FF) (due to early sampling, maternal obesity and some technical factors). Learn more about the causes of false-negative results on NIPT and some of the clinical reports.
CAUSE OF False-Negative NIPT TEST
1. Phenomenon of fetal mosaicism
Placental-placental mosaicism is a challenging biological problem and one of the most frequent causes of discrepancies between NIPT and diagnostic test results. Mosaic accounted for about 20% of all causes of inaccurate NIPT results.
Mosaic is the appearance of two or more cell lines carrying different sets of chromosomes in the same individual. The mosaicism that occurs between the placenta and the fetus can distort the results of the NIPT test. In the case of fetal mosaicism, the fetal cells carry abnormal chromosomes but the placenta carries normal chromosomes, therefore, when performing the NIPT test, analysis of cfDNA released from the placenta could not detect abnormal cells of the fetus.
In the study by Yang et al., 2017 reported a clinical case of a false negative NIPT test.

A 32-year-old woman who became pregnant naturally and had no family history of genetic disease. At 15 weeks' gestation, the NIPT test showed a low risk for trisomy 13, 18, and 21. The woman continued to have routine antenatal checkups, and during the third trimester, ultrasound imaging revealed heart’s abnormalities. After being consulted and prescribed by the doctor, the pregnant woman decided to have amniocentesis to do the karyotype, the results showed that two different cell lines appeared in the fetus: 47,XX,+18[61]/46,XX [39]. In the end, the pregnant woman decided to terminate the pregnancy.
After the termination of pregnancy, QF-PCR and karyotype tests were continued at different sites on the placenta and fetal cord blood to determine the exact cause of this deviation. The results showed that the placenta showed a low risk for trisomy 13, 18 and 21, while cord blood and DNA from amniotic fluid cells showed different cell lines of 47,XX,+18. 61]/46,XX[39]. Conclusion: The occurrence of fetal mosaicism leads to false negative NIPT results.
Currently, technical limitations as well as problems on biological basis do not allow accurate determination of mosaic ratio and type of mosaic through NIPT test. However, understanding the risk of falsifying results that may result from the presence of mosaicism, especially in the case of false negatives, will help pregnant women and doctors be more careful in managing pregnancies, such as the third trimester. second and third trimester, through ultrasound to monitor fetal growth as well as other abnormalities that may appear.
2. Low Fetal Fraction (FF)
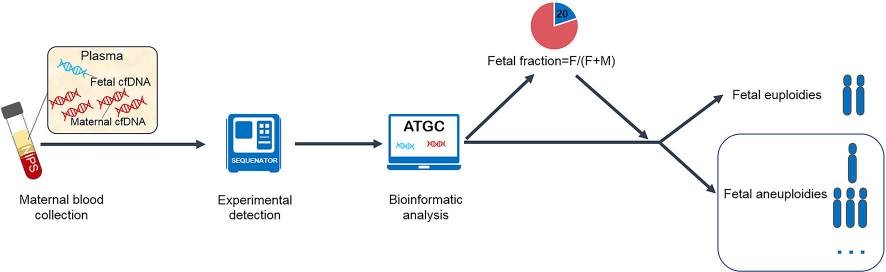
Fetal fraction (FF) is the ratio of fetal cfDNA to all circulating cfDNA in maternal plasma. The NIPT test is based on the analysis of fetal cfDNA released into the maternal blood circulation, therefore, FF is directly related to the ability to detect fetal chromosomal anomalies. When the amount of fetal cfDNA is too low, resulting in a low FF, then it is possible for NIPT results to be falsely negative or unreadable.
Low FF is responsible for up to 50% of false negatives. Factors that can affect the fetal fraction were also investigated and evaluated, including: sampling at an early stage, maternal obesity, and technical problems with testing. also affect a certain percentage.
 2.1. Low FF due to early gestational week sampling
2.1. Low FF due to early gestational week sampling
Fetal cfDNA released into the maternal circulation is reported to be detectable from 5 weeks gestation, from 10 to 21 weeks gestation, FF ranges from 10-15%, in which fetal cfDNA increases 0.1% per week (p < 0.0001). After 21 weeks of gestation, fetal cfDNA increased by 1% per week (p < 0.0001). Fetal cfDNA fragments are approximately 150-160 bp in length, smaller than maternal DNA and account for 11-13.4% of the total cfDNA present in maternal blood. Research results show that the amount of cfDNA of the fetus increases with gestational age and will be lost within a few hours after birth.
A summary report on the rate of FF to ensure the quality of NIPT testing is FF ~ 4%, and the recommended gestational week is from the 10th week of pregnancy. Therefore, when performing maternal blood sampling at gestational weeks earlier than 10 weeks, may result in insufficient fetal cfDNA to read results or lead to inaccurate results.
In the world, the Fertility Associations have also given very specific guidelines on the use of NIPT, NIPT is recommended to be performed from the 10th week of pregnancy and in Vietnam, according to Decision 1807 issued 2020 clearly states: "The recommended week of pregnancy for NIPT testing is from 10 weeks, after having fetal ultrasound results and consulting a genetic counselor for appropriate indications".
2.2. Maternal obesity
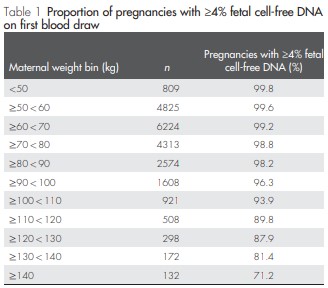
In the 2013 study by Wang et al., the team found a correlation between maternal body mass index (BMI) and total cfDNA. In thin pregnant women, total cfDNA concentrations were shown to decrease due to the dilution effect of increased plasma volume. In contrast, in obese pregnant women, total cfDNA levels were increased due to increased adipose tissue necrosis and increased vascular stromal cell death, which offset the dilution effect. Although fetal cfDNA was not affected, increased maternal total cfDNA resulted in lower FF overall (Table 1).
An increase in maternal body weight or BMI may increase maternal circulating cfDNA concentrations leading to a lower FF ratio. After explaining this phenomenon, many researchers have conducted many regression analyzes to quantify the ratio of FF and BMI in mothers, thereby eliminating the association bias caused by gestational age and BMI.
2.3. Technical problems
In addition to the above main reasons, technical issues such as cfDNA extraction technology, library creation and sequencing, and bioinformatics software reading results are also a factor affecting the accuracy of the results. Up to now, scientists and research centers around the world have always made great efforts to overcome the erroneous rates of NIPT results through the application of new technologies and their incorporation into clinical practice.
NIPT testing at GENTIS uses next-generation sequencing technology on the NextSeq 550 sequencer system of Illumina, USA. Illumina is a leading technology company in the world in gene sequencing, with outstanding quality voted by the worldwide community of scientists as a technology company used in scientific research, pathology, develops tests in the field of molecular biology the most.

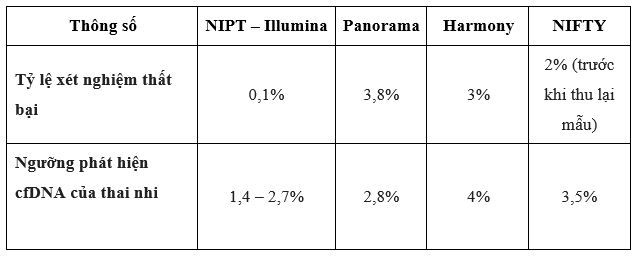
When comparing Illumina's technology platform with some other technology platforms, Illumina shows outstanding advantages in terms of cfDNA detection at low concentrations of 1.4 - 2.7% and test failure rate. (0.1%) is the lowest among the platforms being used today (Table 1). In addition, the GENTIS R&D team has successfully developed a proprietary bioinformatics software that optimizes the readability of the results, thereby improving the accuracy of the NIPT test.
Understanding the causes of unsatisfactory NIPT results is essential for clinicians and genetic counselors to comprehensively advise pregnant women and make appropriate indications both before and after performing the test. Along with choosing a reputable testing unit, using the most advanced and modern technology, it will help doctors and pregnant women get accurate results and clearest advice for more peace of mind. pregnancy management.
References:
[1] Salomon LJ, Sotiriadis A, Wulff CB, Odibo A, Akolekar R. Risk of miscarriage following amniocentesis or chorionic villus sampling: systematic review of literature and updated meta-analysis. Ultrasound Obstet Gynecol. (2019) 54:442–51. 10.1002/uog.20353
[2] Grati FR, Grimi B, Frascoli G, Meco A.M. Di, Liuti R, Milani S, et al. Confirmation of mosaicism and uniparental disomy in amniocytes, after detection of mosaic chromosome abnormalities in chorionic villi. Eur J Hum Genet. (2006) 14:282–8. 10.1038/sj.ejhg.5201564
[3] Mackie F. L., Hemming K., Allen S., Morris R. K., Kilby M. D. (2017). The accuracy of cell-free fetal DNA-based non-invasive prenatal testing in singleton pregnancies: A systematic review and bivariate meta-analysis. BJOG 124, 32–46. 10.1111/1471-0528.14050
[4] Van Opstal, Diane et al. “False Negative NIPT Results: Risk Figures for Chromosomes 13, 18 and 21 Based on Chorionic Villi Results in 5967 Cases and Literature Review.” PloS one vol. 11,1 e0146794. 15 Jan. 2016, doi:10.1371/journal.pone.0146794
[5] Deng C, Liu S. Factors Affecting the Fetal Fraction in Noninvasive Prenatal Screening: A Review. Front Pediatr. 2022;10:812781. Published 2022 Jan 27. doi:10.3389/fped.2022.812781
[6] Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn. 2013;33(7):662-666. doi:10.1002/pd.4119
[7] Yang J, Qi Y, Guo F, et al. A case of placental trisomy 18 mosaicism causing a false negative NIPT result. Mol Cytogenet. 2017;10:40. Published 2017 Oct 27. doi:10.1186/s13039-017-0341-5
[8] Vora, Neeta L et al. “A multifactorial relationship exists between total circulating cell-free DNA levels and maternal BMI.” Prenatal diagnosis vol. 32,9 (2012): 912-4. doi:10.1002/pd.3919
[9] Samura, Osamu, and Aikou Okamoto. “Causes of aberrant non-invasive prenatal testing for aneuploidy: A systematic review.” Taiwanese journal of obstetrics & gynecology vol. 59,1 (2020): 16-20. doi:10.1016/j.tjog.2019.11.003
[10] Illumina. Analytical Validation of the verifi® prenatal test: Enhanced Test Performance for Detecting Trisomies 21, 18, and 13 and the Option for Classification of Sex Chromosome Status. Illumina White Paper. 2012.











