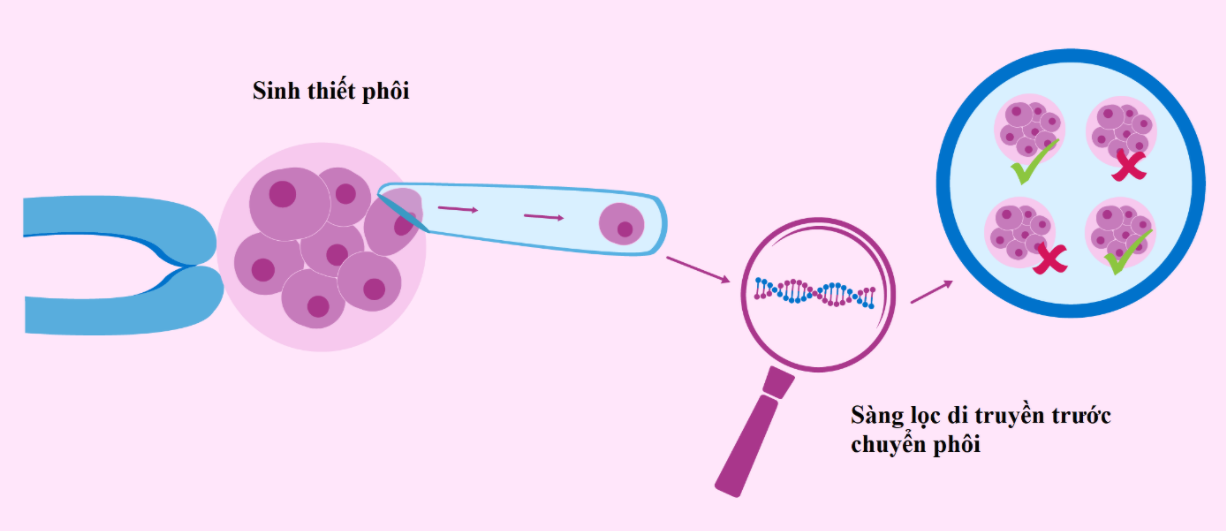
In recent years, the rate of infertility has continuously increased and more and more couples are choosing to use assisted reproductive technology to treat infertility [1]. One of them is the method of in vitro fertilization (IVF) with the most implementation rate. The goal of IVF is to select a good embryo for transfer to the mother's uterus and produce a clinically healthy pregnancy in the shortest possible time.
However, the incidence of chromosomal abnormalities in embryos cultured in vitro can be as high as 60% and this is strongly associated with implantation failure and high miscarriage rates [2]. Therefore, the genetic analysis test before embryo transfer, also known as PGTest (Preimplantation Genetic Testing) was born and marked a big turning point in the field of assisted reproduction. To date, the PGTest test has become a routine method to test for abnormalities related to the number, structure of chromosomes and genetic disorders of monogenic diseases in embryos worldwide.
Technology and application techniques in testing PGTest
The process of performing the PGTest test is quite complicated and requires a team of highly qualified personnel with modern facilities and equipment. Since its inception, the PGTest test has always been updated and optimized in technology to improve screening efficiency. Up to now, with the development of science and technology, many technologies have been developed and used for genetic analysis before embryo transfer such as:
• Fluorescence in situ hydridization (FISH) technique
• Microarray comparative genomic hybridization (aCGH)
• Next generation sequencing (NGS) technology.
Each method has its own advantages and disadvantages, so which method for genetic analysis before embryo transfer has the highest accuracy and the most reasonable cost?
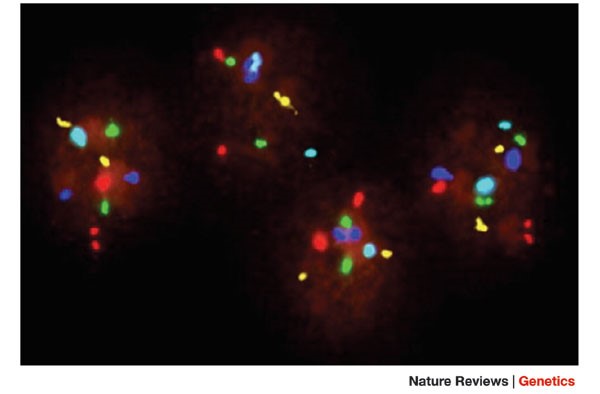
1. FISH - Fluorescence in situ hydridization
In 1995, fluorescence in situ hydridization (FISH) was first applied and considered the main method for genetic analysis before embryo transfer [4].
FISH is a technique of hybridization of fluorescent probes (DNA sequences) with fluorescent target DNA on chromosomes in the form of dividing (metaphase) or non-dividing (interphase) cells. Going through the steps of fixation, denaturation, and hybridization with fluorescein-labeled DNA, the sample will be analyzed under a fluorescence microscope to detect abnormalities in chromosomes and related genes.
However, FISH is limited by the number of fluorescent probes, thus limiting the number of chromosomes tested. Genetic analysis before embryo transfer using FISH only allows testing on five chromosomes, 13, 18, 21, X and Y [5]. In addition, the detection of mosaicism or microdeletion/repeat mutations in embryos is still limited.
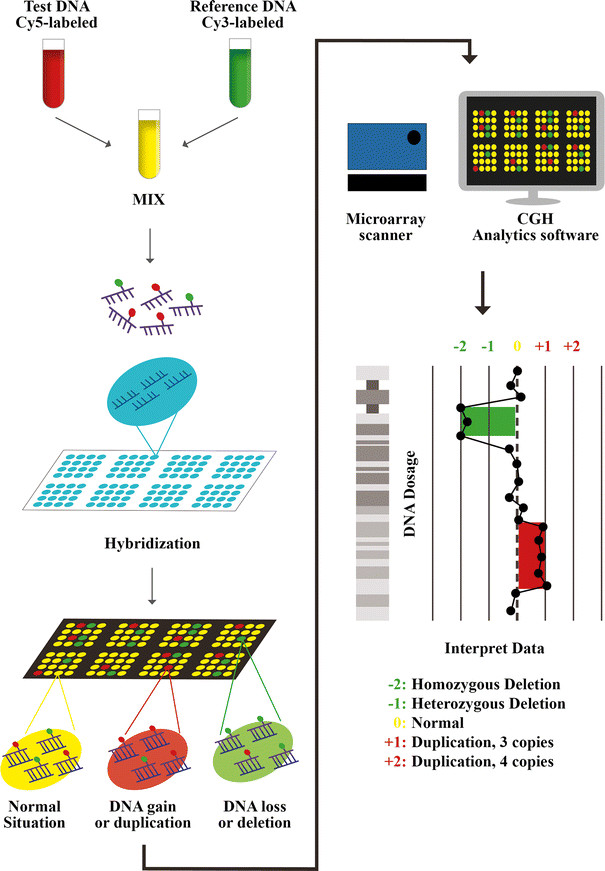
2. Microarray-based comparative genomic hybridization technique
Microarray-based comparative genomic hybridization (aCGH) was applied immediately after FISH. It is a molecular cytogenetic technique to detect genome-wide chromosomal changes with high resolution, aCGH compares a patient's genome with a reference genome and identifies differences. differentiate between the two genomes and thereby identify the region of genomic imbalance in the patient.
This technique was able to detect abnormalities on all 24 chromosomes and mosaicism in embryos, which is far superior to the previous FISH method. However, aCGH is still very limited to detect deletion/repeat mutations or syndromes related to structural abnormalities of chromosomes.
3. New Generation Gene Sequencing (NGS) Technology
This technology is considered the current standard in genetic analysis before embryo transfer. Unlike the above two methods, the NGS approach is based on the direct determination of the nucleic acid sequence of a certain DNA or cDNA molecule. NGS helps to deal with larger-scale datasets, at a much lower recurring cost.
When compared with the above two methods, the researchers showed that:
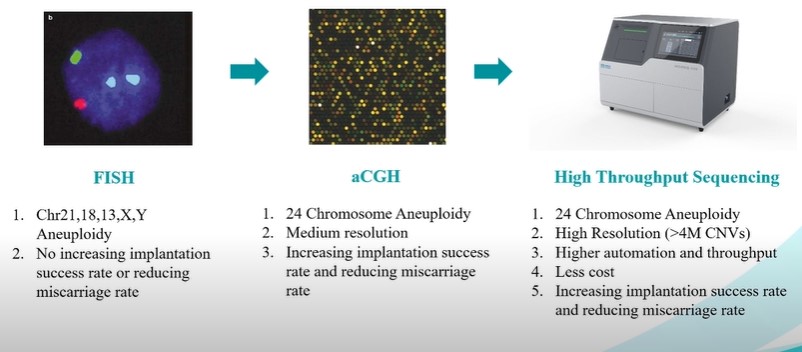
(1) FISH detection of chromosomal abnormalities is much more limited than aCGH and NGS, because FISH only detects abnormalities on five chromosomes 13, 18, 21, X and Y while aCGH and NGS can detect chromosomal abnormalities on all 24 chromosomes [6];
(2) Using next generation gene sequencing (NGS) technology improves the survival rate related to the ability to detect abnormal embryos better than FISH and aCGH technology [7].
From that, it can be seen that the application of NGS technology in genetic analysis testing before embryo transfer has improved and improved the efficiency of screening for abnormalities in embryos, helping to increase the rate of conception and birth of healthy babies. strong.
Next Generation Sequencing – NGS also has many different platforms, including: Illumina sequencing, Proton/PGM (Ion Torrent) sequencing and PacBio RS II sequencing. Each technology has its own advantages and disadvantages, studies around the world have also evaluated the effectiveness of each technology platform when applied in genetic analysis before embryo transfer. The common advantages of the new generation gene sequencing platforms are the analysis of the entire 24 chromosomes, the detection of microdeletion/duplication mutations and a number of syndromes related to structural abnormalities of chromosomes.
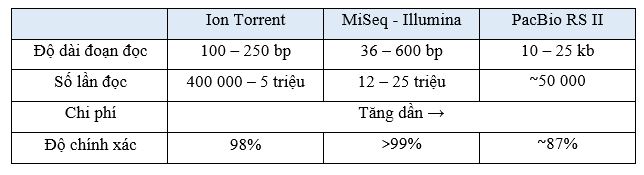
Because the Ion Torrent sequencing platform uses semiconductor technology, not optical technology to measure signals, the time to read and analyze results is fast, and the system is compact. But the limitation of Ion Torrent is that the read length is only about 100 - 250 bp, so at the bases behind the readability will gradually decrease and it is difficult to accurately count the continuous repeating sequence of a nucleotide. Although the machine system is compact, the implementation process is relatively complicated and includes many steps, which is also a cause of errors from many stages of the testing process [8].
As for Illumina's technology platform, the outstanding advantage is in the ability to analyze sequences of a continuously repeating nucleotide and with a read length of 36 - 600 bp, which is larger than that of Ion Torrent. In addition, llumina's technology platform is also proven to have large reading coverage, increasing the readability of the results in the most optimal way.
According to the 2020 statistics, Illumina's technology platform is now more commonly used by the world's scientific community than the Ion Torrent technology platform. However, Illumina sequencing has the disadvantage of longer data analysis time and higher cost compared to Ion Torrent technology.
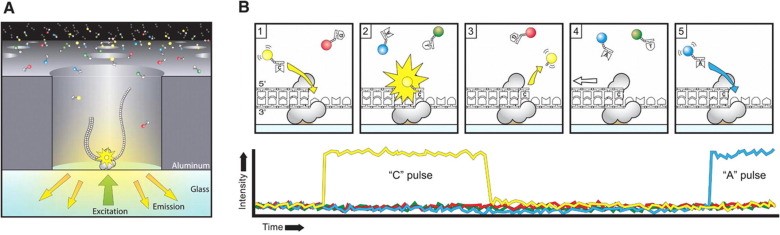
PacBio RS II's technology platform is developed following Illumina's technology platform, with the advantage of large read length, about 10-15 kb with significantly faster result analysis time than Ion Torrent and Illumina. However, PacBio RS II sequencing has a low throughput, PacBio RS II reads are only about 50,000 per run, so the error rate is still high, and the test cost is also higher. compared to the two platforms Ion Torrent and Illumina [9]
With new generation gene sequencing (NGS) technology platforms, outstanding advantages are in depth and resolution, the ability to screen and detect abnormalities with high accuracy. In particular, Illumina's technology platform is considered a trend with the advantages of deeper coverage analysis, so the accuracy is also higher, the price is reasonable, the applicability is wide, and it is considered a standard in genetic analysis before embryo transfer.
Technological foundation of PGTest testing at GENTIS
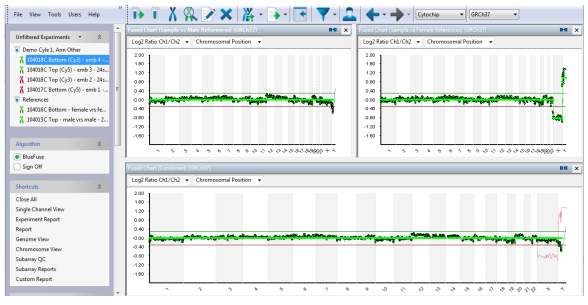
The technology platform used in this assay is Illumina sequencing on the MiSeq system - Illumina, USA, and the analysis software BlueFuse Multi has been shown to have a sensitivity and specificity of up to 99%. With the advantages of the Illumina technology platform such as: the ability to analyze deep coverage, read consecutive repeat sequences of a nucleotide, the PGTesst test at GENTIS helps to increase the rate of selecting high-quality embryos for testing. transferred to the mother's uterus, thereby increasing the rate of successful clinical pregnancy when performing in vitro fertilization (IVF).

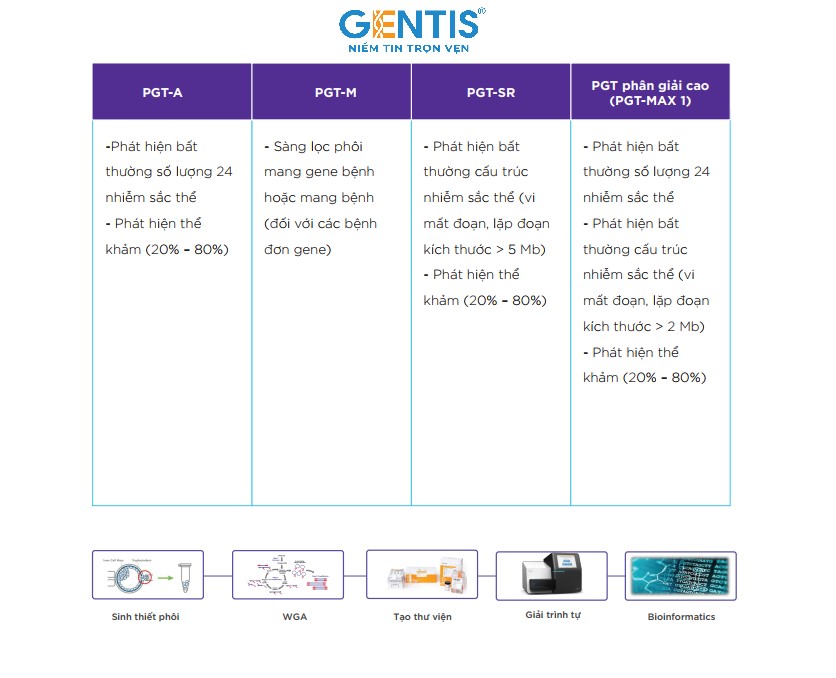
GENTIS is offering 3 pre-transfer genetic testing packages PGT-A, PGT-SR and PGT-M to help select embryos without chromosomal abnormalities, supporting pre-optimized embryo selection. when transferred into the mother's uterus. In addition, GENTIS also launched the PGT Max 1 test package - a breakthrough test from PGTest, exclusively improved with outstanding advantages that no other unit in Vietnam has been able to perform.
The special feature of this test is that in addition to detecting quantitative and structural abnormalities on all 24 chromosomes and common syndromes caused by mutations >5Mb in size, PGT Max 1 can also detected 2 more common microdeletion abnormalities with small size of only 2Mb including 22q11.2 deletion (associated with DiGeorge syndrome) and 1p36 deletion (associated with 1p36 deletion syndrome).
At GENTIS, we are constantly trying to improve service quality, applying the most modern and advanced testing technologies and techniques to bring customers the most comprehensive solutions, contributing to improving the quality of services. effective care and protection of human health.
References:
[1] Vander Borght M., Wyns C. Fertility and infertility: definition and epidemiology. ClinicalBiochemistry . 2018;62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012.
[2] Franasiak J. M., Forman E. J., Hong K. H., et al. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertility and Sterility . 2014;101(3):656–663. doi: 10.1016/j.fertnstert.2013.11.004.
[3] Handyside, A H et al. “Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification.” Nature vol. 344,6268 (1990): 768-70. doi:10.1038/344768a0
[4] Griffin D.K., Ogur C. Chromosomal analysis in IVF: Just how useful is it? Reproduction. 2018;156:F29–F50. doi: 10.1530/REP-17-0683.
[5] Rius M., Obradors A., Daina G., Ramos L., Pujol A., Martínez-Passarell O. Detection of unbalanced chromosome segregations in preimplantation genetic diagnosis of translocations by short comparative genomic hybridization. Fertil Steril. 2011;96:134–142.
[6] Cornelisse, Simone et al. “Preimplantation genetic testing for aneuploidies (abnormal number of chromosomes) in in vitro fertilisation.” The Cochrane database of systematic reviews vol. 9,9 CD005291. 8 Sep. 2020, doi:10.1002/14651858.CD005291.pub3
[7] Bartels, Chantal B et al. “In vitro fertilization outcomes after preimplantation genetic testing for chromosomal structural rearrangements comparing fluorescence in-situ hybridization, microarray comparative genomic hybridization, and next-generation sequencing.” F&S reports vol. 1,3 249-256. 25 Sep. 2020, doi:10.1016/j.xfre.2020.09.011
[8] Lahens, Nicholas F et al. “A comparison of Illumina and Ion Torrent sequencing platforms in the context of differential gene expression.” BMC genomics vol. 18,1 602. 10 Aug. 2017, doi:10.1186/s12864-017-4011-0
[9] Rhoads, Anthony, and Kin Fai Au. “PacBio Sequencing and Its Applications.” Genomics, proteomics & bioinformatics vol. 13,5 (2015): 278-89. doi:10.1016/j.gpb.2015.08.002











